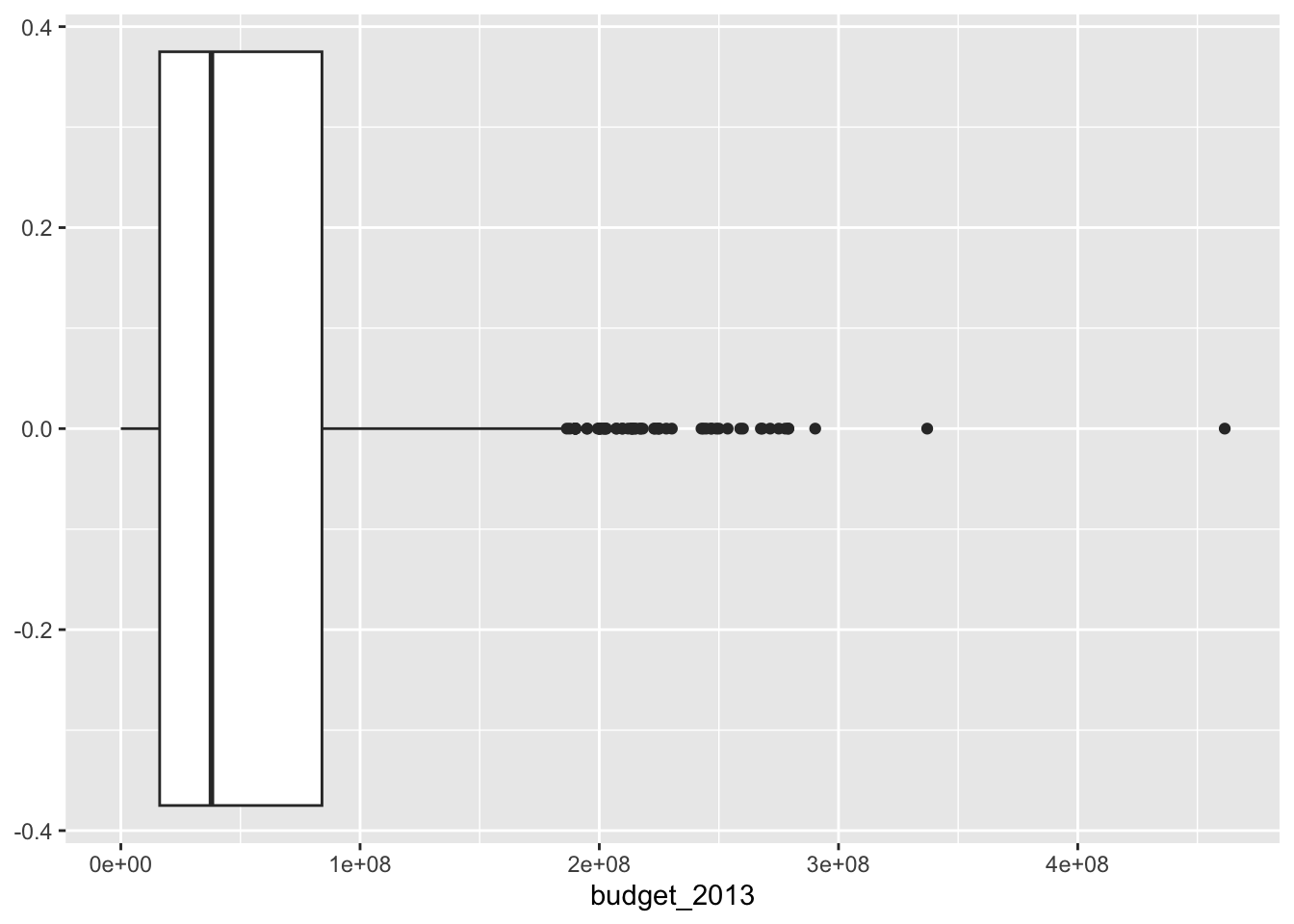
bechdel |>
group_by(binary) |>
summarise(mean_roi = mean(roi, na.rm = TRUE),
mean_budget = mean(budget_2013)) # A tibble: 2 × 3
binary mean_roi mean_budget
<chr> <dbl> <dbl>
1 FAIL 8.36 65877024.
2 PASS 7.99 46913086.